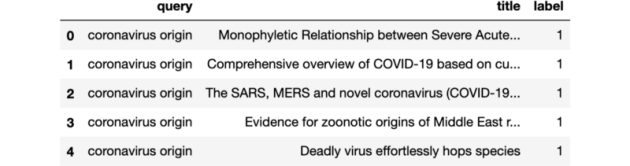
How to ensure training and serving encoding compatibility
There are cases where the inputs to your Transformer model are pairs of sentences, but you want to process each sentence of the pair at different times due to your application’s nature. Search applications are one example.

The search use case
Search applications involve a large collection of documents that can be pre-processed and stored before a search action is required. On the other hand, a query triggers a search action, and we can only process it in real-time. Search apps’ goal is to return the most relevant documents to the query as quickly as possible. By applying the tokenizer to the documents as soon as we feed them to the application, we only need to tokenize the query when a search action is required, saving us precious time.
In addition to applying the tokenizer at different times, you also want to retain adequate control about encoding your pair of sentences. For search, you might want to have a joint input vector of length 128 where the query, which is usually smaller than the document, contributes with 32 tokens while the document can take up to 96 tokens.
Training and serving compatibility
When training a Transformer model for search, you want to ensure that the training data will follow the same pattern used by the search engine serving the final model. I have written a blog post on how to get started with BERT model fine-tuning using the transformer library. This piece will adapt the training routine with a custom encoding based on two separate tokenizers to reproduce how a Vespa application would serve the model once deployed.
Create independent BERT encodings
The only change required is simple but essential. In my previous post, we discussed the vanilla case where we applied the tokenizer directly to the pairs of queries and documents.
from transformers import BertTokenizerFast
model_name = "google/bert_uncased_L-4_H-512_A-8"
tokenizer = BertTokenizerFast.from_pretrained(model_name)
train_encodings = tokenizer(
train_queries, train_docs, truncation=True,
padding='max_length', max_length=128
)
val_encodings = tokenizer(
val_queries, val_docs, truncation=True,
padding='max_length', max_length=128
)In the search case, we create the create_bert_encodings function that will apply two different tokenizers, one for the query and the other for the document. In addition to allowing for different query and document max_length, we also need to set add_special_tokens=False and not use padding as those need to be included by our custom code when joining the tokens generated by the tokenizer.
def create_bert_encodings(
queries, docs, tokenizer, query_input_size, doc_input_size
):
queries_encodings = tokenizer(
queries, truncation=True,
max_length=query_input_size-2, add_special_tokens=False
)
docs_encodings = tokenizer(
docs, truncation=True,
max_length=doc_input_size-1, add_special_tokens=False
)
TOKEN_NONE=0
TOKEN_CLS=101
TOKEN_SEP=102
input_ids = []
token_type_ids = []
attention_mask = []
for query_input_ids, doc_input_ids in zip(
queries_encodings["input_ids"], docs_encodings["input_ids"]
):
# create input id
input_id = [TOKEN_CLS] + query_input_ids + [TOKEN_SEP] +
doc_input_ids + [TOKEN_SEP]
number_tokens = len(input_id)
padding_length = max(128 - number_tokens, 0)
input_id = input_id + [TOKEN_NONE] * padding_length
input_ids.append(input_id)
# create token id
token_type_id = [0] *
len([TOKEN_CLS] + query_input_ids + [TOKEN_SEP]) +
[1] * len(doc_input_ids + [TOKEN_SEP]) +
[TOKEN_NONE] * padding_length
token_type_ids.append(token_type_id)
# create attention_mask
attention_mask.append(
[1] * number_tokens + [TOKEN_NONE] * padding_length
)
encodings = {
"input_ids": input_ids,
"token_type_ids": token_type_ids,
"attention_mask": attention_mask
}
return encodingsWe then create the train_encodings and val_encodings required by the training routine. Everything else on the training routine works just the same.
from transformers import BertTokenizerFast
model_name = "google/bert_uncased_L-4_H-512_A-8"
tokenizer = BertTokenizerFast.from_pretrained(model_name)
train_encodings = create_bert_encodings(
queries=train_queries,
docs=train_docs,
tokenizer=tokenizer,
query_input_size=32,
doc_input_size=96
)
val_encodings = create_bert_encodings(
queries=val_queries,
docs=val_docs,
tokenizer=tokenizer,
query_input_size=32,
doc_input_size=96
)Conclusion and future work
Training a model to deploy in a search application require us to ensure that the training encodings are compatible with encodings used at serving time. We generate document encodings offline when feeding the documents to the search engine while creating query encoding at run-time upon arrival of the query. It is often relevant to use different maximum lengths for queries and documents, and other possible configurations.

We showed how to customize BERT model encodings to ensure this training and serving compatibility. However, a better approach is to build tools that bridge the gap between training and serving by allowing users to request training data that respects by default the encodings used when serving the model. pyvespa will include such integration to make it easier for Vespa users to train BERT models without having to adjust the encoding generation manually as we did above.